DNA Helix Bender Tutorial
A tutorial was added for using the DNA helix bundle bender script https://github.com/dfu99/dnahbbender.
While this only covers a very narrow slice of the design space, it’s useful for what it does cover, which is any continuous, bent helix bundle 2D structure.
The tutorial walks through building a triangle as an example of the type of structures the script can facilitate design for.
Tutorial
This will walk through the design of a 6 helix bundle triangle with continuous, rounded corners.
First, of all, we need to know our exact dimensions before creating a template.
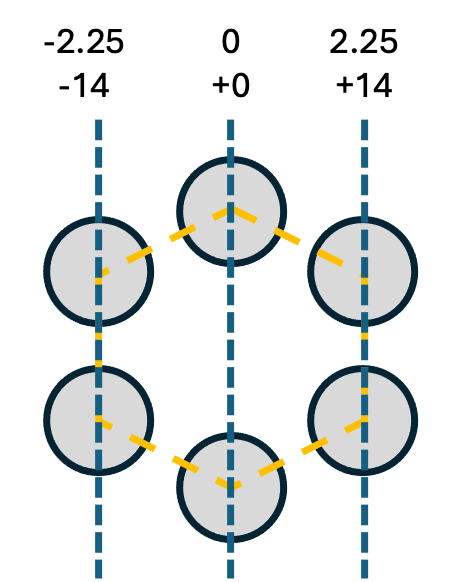
In this design, we’ll be folding a 6 helix bundle along its 3-plane axis. We can mark the planes as such:
We then have to decide how much we want our corners to curve.
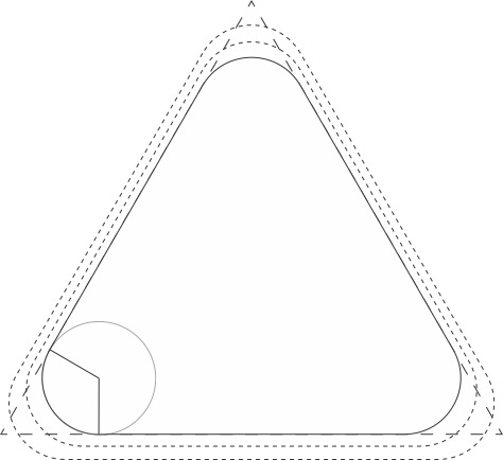
In this case, we are only interested in the inner dimensions of the triangle, but the other layers have been drawn with dotted lines. The longer dashed line at the triangle’s corners are how much we will be shortening an edge to insert the curved corner. We started with a 126 bp edge.
For this structure, with a bit of trig, we determined that, along our bend, our inner layer should have 32 bp (approximately 3 full turns).
Using the beam model equation above, for the positions of our three planes, the insertion/deletion gradient is [-14, 0, +14]. Thus, with 32 bp as our reference, the subsequently outer layer bends are 46 bp and 60 bp long.
We use the middle plane length as the nominal length for creating the caDNAno template. The total length of our triangle (as a cycle) is \(46 \cdot 3 + 84 \cdot 3 = 390\).
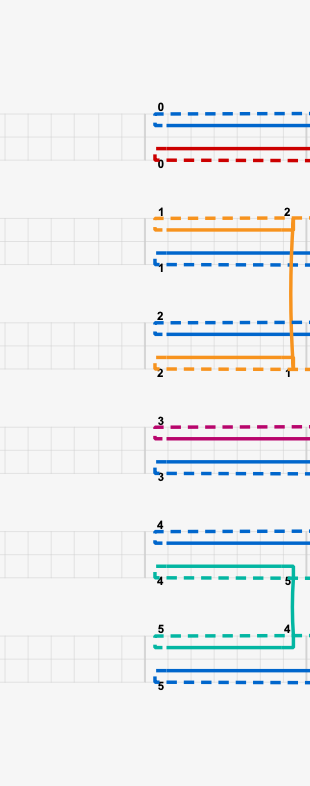
We then go on and draw that in caDNAno.

Add our breakpoints…

Note that the endpoints of the structure are connected.
 - … -
- … - 
We now run the script, three times, once to make each corner.
python autobender.py -o temp_01.json -lt hc -a 120 -l 46 -s 91 -x 0 template.json
Produces…

For the second bend…
python autobender.py -o temp_02.json -lt hc -a 120 -l 46 -s 221 -x 0 temp_01.json

And for the third bend…
python autobender.py -o temp_03.json -lt hc -a 120 -l 46 -s 351 -x 0 temp_02.json

This final caDNAno design can then be converted by tacoxDNA and simulated in oxDNA or viewed in oxView.
