DNA Complexes Dashboard
I’ve been working on a DNA foundation model, not for genomics, which seems to be the usual lately, but for primarily DNA computing. There’s a subtle thread throughout the years of this idea that DNA computing can probably do much more than what’s been demomnstrated. It struggles to reach its full potential in designing complex, dynamical systems because we’re a bit strapped behind having to design obvious reactions. We can’t make much sense of the less than obvious systems.
Strategies to circumvent this bottleneck are few but quite interesting. The theme of being able to distinguish the full spectrum orthogonal and non-specific hybridization reactions is likely a pivotal idea 1,2,3. Use and understand mismatches, rather than avoid them completely.
In any case, I’m quite invested in seeing how such a philosophy develops.
I’m sharing a useful widget here that’s come in handy for analyzing some smaller systems. I generated some synthetic training data using NUPACK to feed into a model. I was plotting inputs side-by-side for a bit, but I couldn’t get a feel for how systems were changing.
This lets me scroll through each data point. I can also select specific complexes and as I scroll, only those complexes (or sums of them!) will be displayed.
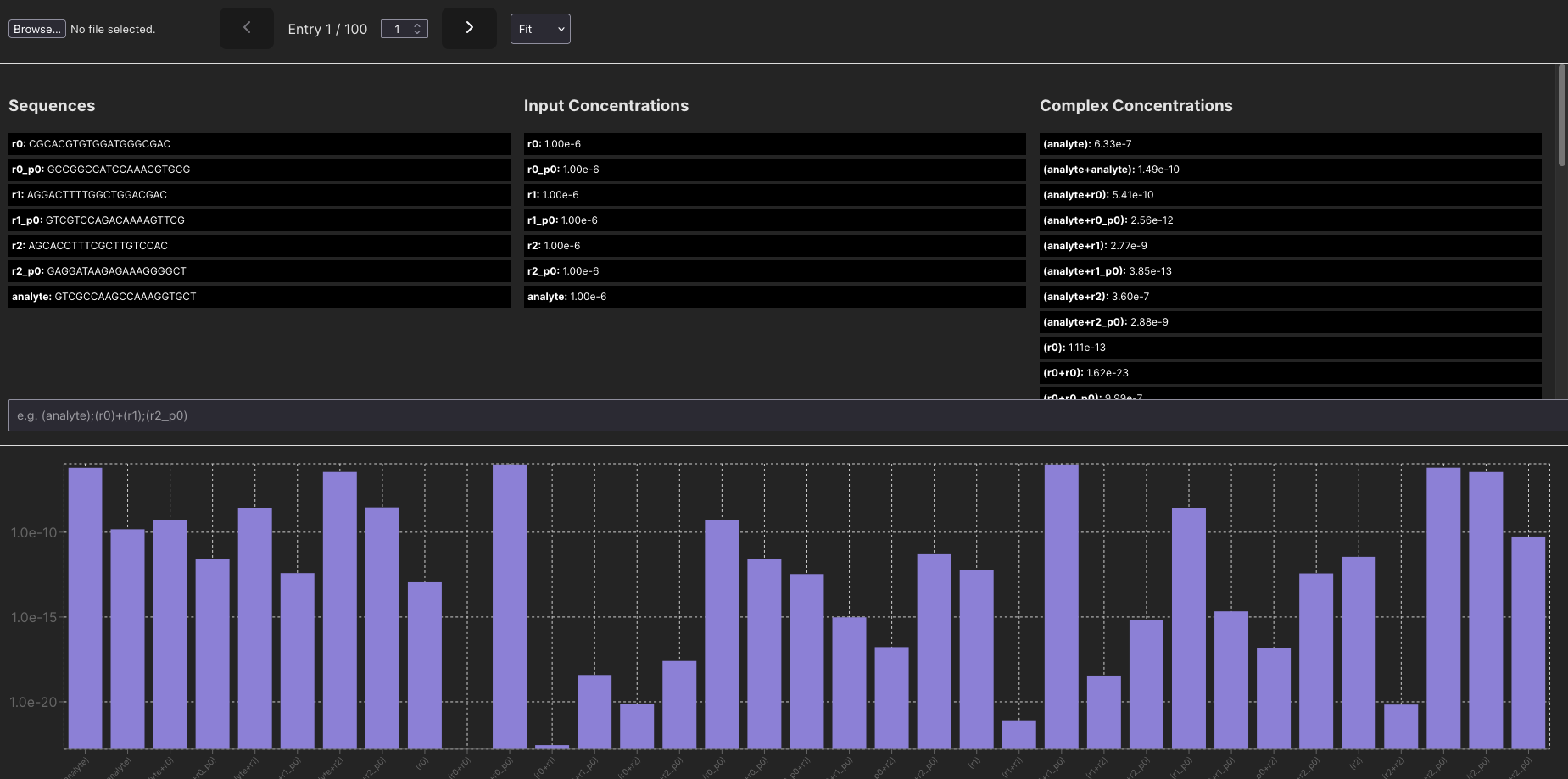 What the interface looks like right now
What the interface looks like right now
This is a first release. Chances are it will continue to change based on whatever my next frustration with handling the data is!
Check it out: https://github.com/dfu99/dna-complex-dashboard
-
Buterez, David. “Scaling up DNA digital data storage by efficiently predicting DNA hybridisation using deep learning.” Scientific reports 11.1 (2021): 20517. ↩
-
Nikitin, Maxim P. “Non-complementary strand commutation as a fundamental alternative for information processing by DNA and gene regulation.” Nature Chemistry 15.1 (2023): 70-82. ↩
-
Su, Yanqing, et al. “Nano scale instance-based learning using non-specific hybridization of DNA sequences.” Communications Engineering 2.1 (2023): 87. ↩