Projects
FIND-SNP
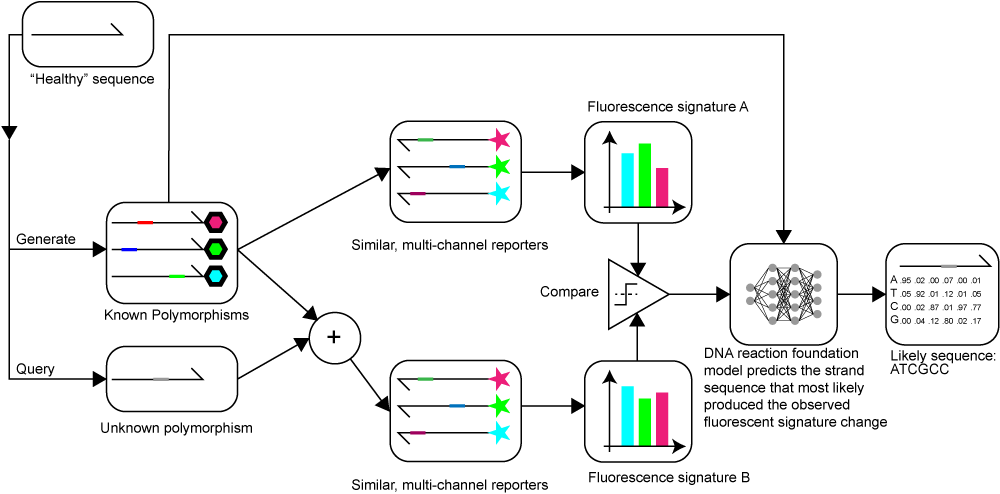
FIND-SNP is a diagnostic framework that leverages deep learning and multi-channel fluorescent assays to identify and locate cancer-related polymorphisms, offering a cost-effective alternative to traditional sequencing methods.
This project is active and ongoing.
Random dendritic ANN generator
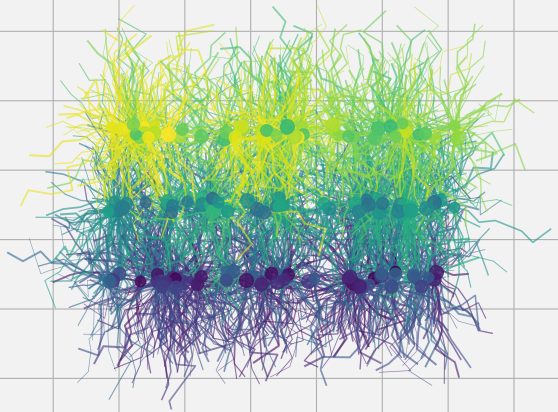
We are creating digital models of brain-like neural structures and transforming them into deep neural networks. By simulating biological growth processes, we can manipulate these structures and study how connectivity shapes learning and performance. This approach helps us uncover principles of neural organization and design more efficient AI models.
This project is active and ongoing.
Hallu-RL
Graphical abstract coming soon
We are developing an AI system where LLMs teach each other in an automated teacher-student-examiner setup. By leveraging controlled hallucinations and reinforcement learning, our approach allows AI agents to refine their understanding of domain-specific knowledge—like financial reports—beyond their pre-training. This presents a new way to view AI-generated speculation, not as an error, but as a tool for deeper learning. We envision that such a framework will also allow us to explore modes by which AI agents can communicate and “copy” their information.
This project is active and ongoing.
DNAxiS
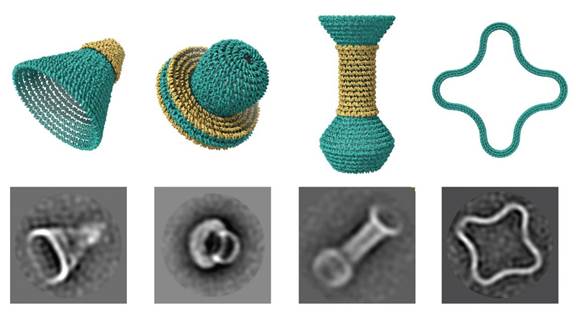
DNAxiS is a computational tool for generating design sequences for rotational DNA origami nanostructures.
You can find it kindly hosted by the Duke University Computer Science Department, here: caddna.cs.duke.edu
It can also be run on your local machine and the source is available on GitHub: https://github.com/dfu99/dnaxis
Our paper on this work was published in Science Advances: DOI: 10.1126/sciadv.ade4455.
Y-STAM
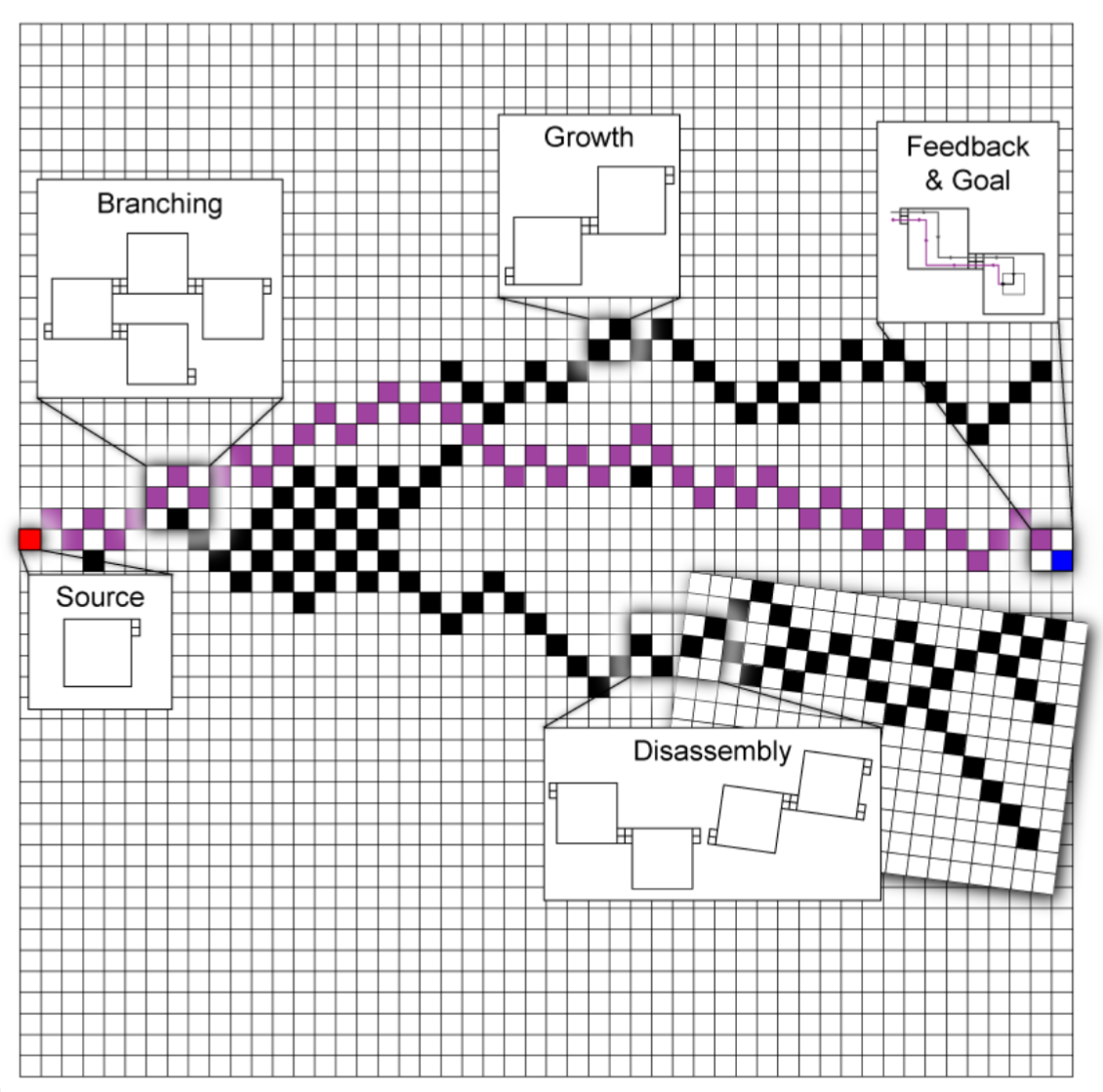
The Branching Signal-passing Tile Assembly Model (Y-STAM) is a model of branching self-assembly processes evaluated using DNA tile assembly as a substrate. It is currently presented as a simulator implemented in MATLAB.
The source code for the MATLAB simulation software is available on GitHub: https://github.com/dfu99/Y-STAM
Our paper on this work was published in the Journal of Royal Society Interface: DOI: 10.1098/rsif.2023.0755
Applications of Localized Hybridization Chain Reaction Circuits
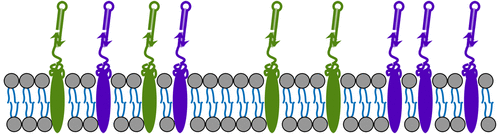
We explored applications of localized hairpin-based HCR circuits [1]. We published work on using our architecture for cancer cell detection in JACS: DOI: 10.1021/jacs.9b05598.
Other
- Krashen-based “i+1” GPT-powered language learning app[GitHub]
- Cambridge dictionary scraper for creating English-Bilingual vocabulary practice worksheets [GitHub]
- 2015 UBC BIOMOD Undergraduate Student Team [Project Website]